How much HIV is in SARS-CoV-2? Fauci's 2018 patent, the NEW Synthetic SARS-CoV-2 Spike gene, with the OLD content.
URGENT: After sending out this post all email updates regarding it are NON-existend. Must have been the substack server or ‘something else’, and now it runs fine :) How surprising the Micro-soft products can be..
Btw., if the real Dr. Fauci’s name would be Dr. fAuIC, now read backwards the capital letters, then probably we could explain the incredible amount of tortures in all the experiments he participated in…
EXPERIMENTAL GENE MODIFICATIONS OF HUMANITY in EU, reactions of the few in parlament:
Update 10/29/2022: Prof. S. Bhakdi from Germany explains with simple words some of the details of immune response and resulting brain and heart damage after covid gene modifying injections: https://usawatchdog.com/cv19-vax-destroys-hearts-brains-of-billions-of-people-dr-sucharit-bhakdi/
Whoever can, please keep Prof. Bhakdi in your prayers. He is facing 5 years in prison and loosing his pension, all for speaking out the truth about covid injections! That’s Germany in 2022!!!
Update 12/14/2022 A link to Mod-E-RNA future plans, including their AI academy, AFTER ruining billions of HUMANS with their genetically modifying covid injections, at: https://s29.q4cdn.com/435878511/files/doc_presentations/2022/11/Moderna-Final-ESG-Day-Slides-11.10.22.pdf was send out by :
Update 12/9/2023: Dr. David Martin uncoveres increidble important facts connecting AZT. HIV and SARS-CoV-2 : https://www.bitchute.com/video/MMXfRHgFybqK/
Clarification.
All my posts talking about the SARS-CoV-2 Spike protein, always refer to the NIH publicly accessible data base for the first OFFICIAL SARS-CoV-2 version at: https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2 with the code for the Spike: YP_009724390.1
The reason for that is the fact, that all the first covid19 injections, Pfizer and ModE-RNA in particular, should OFFICIALLY contain genetic material encoding the Spike described in these data bases. Any conclusions drawn from all the bioinformatics analysis, refer thus to facts about that genetic material of covid ILLEGAL genetically modifying injections.
The POST starts below, but if you read the above, that’s ok too, it will help in the long run:) THANK YOU for continuing:) And this post like others too, will be updated and changed when new insights flow in.
For the last ~ 3 YEARS Dr. Fauci is flattening the curve, which at the begin of 2020 was not even there. He flattened many things in the meantime and that while for many, it almost looks like ‘fortune cookie game"‘ is the main feature of this man. Many think he does not know anything, including the late RIP Dr. Kary Mullis, who had to die right before the pandemic, and who also not only was the top specialist on the PCR technology, used so reckless during the covid pandemic, but who was also one of the first doing an extensive research on ACE receptors and alveolar hypoxia^8.
So does Fauci know ‘things’ or he does not? Whoever made the below caricature, THANK YOU.
Fact check: “SARS-CoV-2 was not created using genes from HIV, Fauci does not hold patents for an ‘HIV component’ to SARS-CoV-2. “
That comes from: https://www.reuters.com/article/uk-factcheck-hiv-covid-explained-idUSKBN29C26E
It’s hard to find any paper which would show how similar HIV gp160 actually is in comparison with the original Spike2020 (my own definition used bellow), from the original SARS-CoV-2 (ncbi entry: /protein_id="YP_009724390.1"). The one which was written by the indian team quickly disappeared from the horizon. Given that the 2008 Walensky’s patent from years back had already quite large pieces of the Spike2020 one ~100% of it and was officially intended to be for HIV treatment, including coronaviruses, how about US 9,896,509 B2 from Feb 2018 with Fauci as one of the inventors? The title of it is:”USE OF ANTAGONISTS OF THE INTERACTION BETWEEN HIV GP120 AND a4b7 INTEGRIN”.
Blast comparison of HIV-gp160 (gp160=gp120+gp41, its amino acid sequence in Query line, available at https://www.ncbi.nlm.nih.gov/protein/9629363 ) with Spike protein (Sbjct line), with the middle line showing the identical amino acids common for both proteins, gives the following fragments, starting with the most tryptophan rich sections:
Query 6 KYQHLWRWGWR-WGTMLLGMLMICSAT 31 >>>HIV-1 gp160
KY+ +W W W + G++ I+ T >>>IDENTITIES
Sbjct 1205 KYEQYIKWPWYIWLGFIAGLIAIVMVT 1231>>>Spike2020
or extending it more
Query 6 KYQHLWRWGW-RW-----GTMLLGM--LMICSAT 31 >>>HIV-1 gp160
KY++ +W W W G + + M +M+C T >>>IDENTITIES
Sbjct 1205 KYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMT 1238 >>>Spike2020
Query 386 NSTQ-LF-----NSTWFNSTWSTEGSNNTEGSDTITLP 417>>>HIV-1 gp160
+STQ LF N TWF++ + G+N T D LP >>>IDENTITIES
Sbjct 49 HSTQDLFLPFFSNVTWFHAIHVS-GTNGTKRFDNPVLP 85 >>>Spike2020
here almost 100 residues in one string:
Query 158 SFNISTSIRGKVQKEYAFFYKLDIIPIDNDT-TS-YKL--TSCN-----------TSVIT
S N+ + + G Y F K ++ P + D+ T Y+ T CN S
Sbjct 438 SNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGF
Query 203 QACPKVSFEPI------------PIHYCAPAGFAIL---KCNNKTFNG 235
Q + V ++P P C P + L KC N FNG
Sbjct 498 QPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNG 545
Query 791 EALKYWWNLLQYWSQELKNSAVSLLNATAIAVAEGTDRVIEVVQ 834
E KY+ N + S ++ +S +NA+ + + + +DR+ EV++
Sbjct 1151 ELDKYFKN---HTSPDVDLGDISGINASVVNIQKEIDRLNEVAK 1191
Query 392 NSTWFNST-WSTEGSNNTEGSDTITLP 417
N TWF ++ + G+N T+ D LP
Sbjct 61 NVTWFHAI-HVS-GTNGTKRFDNPVLP 85
Query 208 VSFEPI--PIHYCAPAGFAIL---KCNNKTFNG 235
+SFE + P C P + L KC N FNG
Sbjct 513 LSFELLHAPATVCGPKKSTNLVKNKCVNFNFNG 545
Query 678 WLWYIKL------------FIMI 688
W WYI L IM+
Sbjct 1212 WPWYIWLGFIAGLIAIVMVTIML 1234
or better
Query 678 WLWYIKL-FIMIVGGLVGLRIV 698
W WYI L FI GL+ + +V
Sbjct 1212 WPWYIWLGFI---AGLIAIVMV 1230
Query 624 NHTTWMEWDREINNYTS 640
N+ +WME E Y+S
Sbjct 148 NNKSWME--SEFRVYSS 162
Query 716 SFQTHLPTPR 725
S+QT+ +PR
Sbjct 673 SYQTQTNSPR 682
Query 652 QQEKNEQELL 661
+Q+KN QE+
Sbjct 773 EQDKNTQEVF 782
Query 458 GGNSNNESEIFR 469
GGN N +FR
Sbjct 446 GGNYNYLYRLFR 457
Query 495 GVAPTK 500
GV+PTK
Sbjct 381 GVSPTK 386
Query 368 DPEIVT--HSF---NC 378
+P+I+T ++F NC
Sbjct 1111 EPQIITTDNTFVSGNC 1126
Query 433 AMYAPPISGQIRCSSNITGLLL 454
+MY I G S+ + LLL
Sbjct 739 TMY---ICGD---STECSNLLL 754
Query 377 NCGGEFFYCNSTQLFNSTWFNS 398
NC ++ + L+NS+ F++
Sbjct 360 NCVADY-----SVLYNSASFST 376
Query 705 VNRVR---QGYSPL-------------SFQTHLPTPR 725
+N VR QG+S L FQT L R
Sbjct 210 INLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHR 246
Query 203 QACPKVSFEPI------------PIHYCAPAGFAIL---KCNNKTFNG 235
Q V ++P P C P + L KC N FNG
Sbjct 498 QPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNG 545
Query 235 GTGPCTNVSTVQCTHGIRPV--VSTQLLL--NGSLAEEEVVIRSVNFTDNAKTIIVQLNT
G+G C++ T Q T+ R V++Q ++ SL E SV +++N+ +I
Sbjct 667 GAGICASYQT-Q-TNSPRRARSVASQSIIAYTMSLGAEN----SVAYSNNSIAIPTNFTI
Query 291 SV--EI 294 HIV-1 gp160
SV EI
Sbjct 721 SVTTEI 726 Spike2020 (furin site start at 681 above)
Query 227 KCNNKTFN----------GTGPCTNV 242
K N K F G+ PC V
Sbjct 458 KSNLKPFERDISTEIYQAGSTPCNGV 483
Query 197 NTS----VITQA--CPKVSFEPIPIH 216
NTS V+ Q C V P+ IH
Sbjct 603 NTSNQVAVLYQDVNCTEV---PVAIH 625
and the comparison below is very special because it contains the homolog of the minimal essential epitope (mentioned in Fauci's patent), a SIMPLE TRIPEPTIDE LDT=Leucine-Aspartic Acid-Threonine(MadCAM), just THREE amino acids, or in case of fibronnectin it is LDV/LDI motif(integrin antagonist), with special function*, given that L,I,V can be substituted due to their molecular equivalence**.
Query 179 LDIIPIDNDTTSYKLTS--CNTS 199 HIV gp-160
LDI P S +++ +NTS IDENTITIES
Sbjct 585 LDITPCSFGGVS--VITPGTNTS 605 Spike2020
* quote from ^1:"Tenascins are extracellular matrix glycoproteins that act both as integrin ligands and as modifiers of fibronectin-integrin interactions to regulate cell adhesion, migration, proliferation and differentiation. In tetrapods, both tenascins and fibronectin bind to integrins via RGD and LDV-type tripeptide motifs found in exposed loops in their fibronectin-type III domains."
** Fauci's patent explains that:
Conservative amino acid substitutions providing functionally similar amino acids are well known in the art. The following six groups each contain amino acids that are conservative substitutions for one another:
1) Alanine (A), Serine (S), Threonine (T);
2) Aspartic acid (D), Glutamic acid (E);
3) Asparagine (N), Glutamine (Q);
4) Arginine (R), Lysine (K);
5) Isoleucine (I), Leucine (L), Methionine (M), Valine (V); and
6) Phenylalanine (F), Tyrosine (Y), Tryptophan (W).
thus conforming: "Antagonists of α4 integrins are known, and include antibodies, peptide antagonists, LDV/LDI peptides, peptidomimetic antagonists, proteomimetic antagonists, and small molecule antagonists."
BLAST output comparing the HIV-gp160 protein amino acid sequence NP_057856.1 with SARS-CoV-2 Spike results in ~89% of the entire sequence coverage of both with ~45% IDENTITIES. If the origin of HIV originated from non-human primates, how is it that the 2 outer proteins, i.e. Spikes coating HIV and the SARS-CoV-2 have so many common amino acid sections??? Two completely different origins of HIV and SARS-CoV-2 can’t end up with so many essential identities, unless the bats landed once in San Francisco and ended up with AIDS, if one believes in the ‘official narrative’! HIV is OLD, SARS-CoV-2 is NEW. Lets take the next toxic HIV tiny TAT protein, transcriptional activator; viral regulatory protein required for virus replication (https://www.ncbi.nlm.nih.gov/protein/9629358 ), protein which shares extremely important 3 common features with Spike2020: RGD motif, very unique cysteine cluster at the C-terminal of Spike, and part of the furin site, here the BLAST output, with Query being the HIV proteins, Subjct line the Spike, and in the middle the amino acids identical for both:
Query 20 TACTNCYCKKCCFHCQVC 37 <<<HIV TAT
T+C C C K C C C <<<IDENTITIES
Sbjct 1238 TSC--CSCLKGCCSCGSC 1253 <<<SPIKE2020
another way to align and extend the homolog section, by hand
C--TNC-YCKK-CCFHCQVC--F HIV TAT
C T C C K CC C C F IDENTITIES
CCMTSCCSCLKGCCS-CGSCCKF Spike2020 SPIKE2020
The same Cys-rich TAT section compared with different Spike piece:
Query 23 TNCYCKKCCFH-CQVCFITKALGISYGRKKR 52 HIV TAT
TN K C F C F LG+ Y + + IDENTITIES
Sbjct 124 TNVVIKVCEFQFCNDPF----LGVYYHKNNK 150 SPIKE2020
Query 46 SYGRKK 51 HIV TAT
++ RK+ IDENTITIES
Sbjct 352 AWNRKR 357 SPIKE2020
this most important section is NOT found by BLAST, here by hand:
SY---GRKKRRQRRRAHQNSQTHQAS HIV TAT
SY RR R A Q + S
SYQTQTNSPRRARSVASQSIIAYTMS Spike2020 FURIN SITE
another section of Spike with the cys-rich
PGSQPKTACTNCYCKKCCFHCQV
P ++ NCY ++ Q
PC-NGVEGF-NCYFPLQSYGFQP
The integrin binding RGD motif by hand, since BLAST 'can't find it':
S-QPRGDPTGPKE HIV TAT
S RGD ++ IDENTITIES
SFVIRGD--EVRQ SPIKE
another section, by hand:
RAHQN-SQTHQASLSKQPT-SQPRG-DPT
RA + Q Q Q + T
RALTGIAVE-QDKNT-QEVFAQVKQIYKTWhat are the HIV TAT’s physiological consequences?
“Neonatal intrahippocampal injection of the HIV-1 proteins gp120
and Tat: Differential effects on behavior and the relationship to
stereological hippocampal measures“^2“HIV-1 Proteins, Tat and gp120, Target the Developing Dopamine System“^3
“HIV-1 proteins gp120 and tat induce the epithelial–mesenchymal transition in oral and genital mucosal epithelial cells“^4
do we have similar issues within SARS infections? Here few citations mentioning similar pathological effects in covid infections:
Ad. 1 “SARS-CoV-2 spike protein induces cognitive deficit and anxiety-like behavior in mouse via non-cell autonomous hippocampal neuronal death“^5
Ad. 2 “SARS-CoV-2 Infection Causes Dopaminergic Neuron Senescence“*6
Ad. 3 “Epithelial-mesenchymal transition induced by SARS-CoV-2 required transcriptional upregulation of Snail“^7
These are just quick searches indicating few common features between HIV and SARS-CoV-2. A far more serious real live report on HIV and covid injections (encoding the Spike and producing it in billions), quite a while ago, by Dr. Nathan Thompson at: https://www.brighteon.com/fa58826c-654f-4d17-bc86-ee86364b2b54
“Dr Nathan Thompson test the blood of his Vaxxed patient!”
Natural Killer T-Cells, CD4, CD8 progression of HIV infection results in blood values which look very similar to the ones AFTER covid genetically modifying injections, NOT infections!!
Please recall, HIV ‘showed up’ in years when there was NO GRAPHENE yet, and yet, the HIV’s spike and SARS-CoV-2 spike are ‘macromolecularly’ RELATED. There is nothing without a purpose in a science paid by the richest!
Now a jump to recent developments. According to an DoD investigation by Tod Callender at:
https://principia-scientific.com/marburg-virus-will-be-activated-in-the-vaxxed-via-5g/
the next ‘coming crisis’ is supposedly related to Marburg virus. Lets’ try the NIH entry for it at: https://www.ncbi.nlm.nih.gov/nuccore/OL702894.1 Is it much different from the above analysis? Keeping all the NIH’s BLAST search parameters identical, targeted for the smaller sections (epitopes connections), the comparison between Marburg glycoprotein UFZ14320.1 and the original SARS-CoV2 Spike glycoprotein shows that they are 62% IDENTICAL, in particular when taking the smaller fragments. Here few examples with Query being Marburg and Sbjct Spike2020:
Query 364 YNTQSTATENEQTSAPSK----TTLLPTENPTTAKSTNSTKSPTT-TVPNTT 410
Y TQ+ + ++ A T L EN + A S NS PT T+ TT
Sbjct 674 YQTQTNSPRRARSVASQSIIAYTMSLGAEN-SVAYSNNSIAIPTNFTISVTT 724
Query 191 LTSTNK-YWTSSNGTQTNDTGC 211 <<<Marburg SPike
LT T + Y T SN QT +GC <<<IDENTITIES
Sbjct 629 LTPTWRVYSTGSNVFQTR-AGC 649 <<<SARS-CoV-2 SPike2020
Query 7 LISIILI 13 <<< Marburg SPike
LI+I+++ <<<IDENTITIES
Sbjct 1224 LIAIVMV 1230 <<<SARS-CoV-2 SPike2020
Query 429 VYFRRKRNILWREGD 443
VY+ K N W E +
Sbjct 143 VYY-HKNNKSWMESE 156
Query 519 WSVQEDDLAAGLSWIPFFGPGIEGLYTAGLI 549 MARBURG
W + W+ F AGLI
Sbjct 1214 WYI----------WLGFI---------AGLI 1225 Spike
Query 318 VVTEPGKTNTTAQ 330
V+T PG TNT+ Q
Sbjct 597 VIT-PG-TNTSNQ 607
Query 242 VKLTSTSTDATELNTTDTNS 261
V LT+ T+L + TNS
Sbjct 16 VNLTTR----TQLPPAYTNS 31
Query 390 PTTA----KSTN 397
P+T KSTN
Sbjct 521 PATVCGPKKSTN 532
Query 226 CAPPKKPSPL 235
C+ PKK + L
Sbjct 525 CG-PKKSTNL 533
Query 300 PQPSTPQQ 307
P PS P +
Sbjct 807 PDPSKPSK 814
Query 141 GAFFLYDRIAST---TMYRGKVF 160
G +F AST + RG +F
Sbjct 89 GVYF-----ASTEKSNIIRGWIF 106
Query 304 TPQQGGNNTNHSQGV-VTEPGKTNTTAQ 330
TP G GV V PG TNT+ Q
Sbjct 588 TPCSFG-------GVSVITPG-TNTSNQ 607
although the tryptophan rich section is at the C-terminal of MarburgVirus glycopprtein, it does not come up within the alignment, so here by hand:
KEGTGWGLGGKW-WTSDWGVLTNL <<< Marburg GLycoprotein
E + KW W W +
QELGKYEQYIKWPWYI-WLGFIAG <<< SARS-CoV-2 Spike
or now shifted section
Query 643 KW-WTSDWGVLTNLGILLLLSIAVLIALSCI-C 673
KW W G + L +++++I + SC C
Sbjct 1211 KWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSC 1243
Query 533 IPFFGPGIEGLYTAGLIKNQNNLVCRLRRLANQT 566
IP G GI Y + Q N R R +A Q+
Sbjct 664 IPI-GAGICASY-----QTQTNSPRRARSVASQS 691 <<<Furin in Spike
Query 255 NTTDTNSDDE-----DLT 267 Marburg glycoprotein
+TTD D + D+T
Sbjct 571 DTTDAVRDPQTLEILDIT 588 <<Spike homolog with Fauci's epitope
note that the non-overlapping segments sometimes are containing exactly the same pool of amino acids, but arranged in different way, frequently backwards, RNA interference???Is this all just random phenomenon??? I DO NOT THINK SO. Molecular biologists learn how to cut and paste pieces of all different organisms, how to change single amino acids to completely change the characteristics of the ‘outcome’. Again and again, every single amino acids counts, a single tripeptide like ‘RGD’ or ‘DRY’ already characterizes the entire feature of a specific protein. If Spike is so similar with other viral proteins, is it also similar with HUMAN proteins?? Almost every post of mine touched that issue, but here once again, some completely random tests on few of them.
A check for homologies with Dipeptidyl peptidase DPP4, membrane glycoprotein (https://www.uniprot.org/uniprotkb/P27487/entry), 92% sc and 37% identities, among others, when searched for smaller fragments:
Query 1 MKTPWKVLLGLLGAAALVTIITVPVVL 27
+K PW + LG + A L+ I+ V + L
Sbjct 1210 IKWPWYIWLGFI--AGLIAIVMVTIML 1234
Query 558 VFRLNWATYLASTE--NII 574
V N Y ASTE NII
Sbjct 83 VLPFNDGVYFASTEKSNII 101
Query 153 QWVTWSPVGHKLAYVW 168
Q++ W P Y+W
Sbjct 1208 QYIKW-PW-----YIW 1217
Query 177 EPNLPSYRITWTGKEDIIYNGIT 199
+P PS R + ED+++N +T
Sbjct 808 DPSKPSKR---SFIEDLLFNKVT 827
or Spike2020 VERSUS human integrin beta3 (https://www.uniprot.org/uniprotkb/P05106/entry) 87% sc, 50% identities(!!):
24.7 bits(50) 0.035 13/26(50%) 16/26(61%) 4/26(15%)
Query 371 VDAYGKIRSK---VELEVRDLPEELS 393 <<<Human integrin beta3
+D Y KI SK + L VRDLP+ S <<<IDENTITIES
Sbjct 197 IDGYFKIYSKHTPINL-VRDLPQGFS 221 <<<Spike2020
Query 593 CMSSNGLLCSG-RGKCECGSC 612
CM+S CS +G C CGSC
Sbjct 1236 CMTS---CCSCLKGCCSCGSC 1253
Query 566 GQCSCGDCLCDSD 578
G CSCG C C D
Sbjct 1246 GCCSCGSC-CKFD 1257
Query 484 GVCRCGPGWLGSQCECSEEDYRP 506
G C C GS C E+D P
Sbjct 1246 GCCSC-----GSCCKFDEDDSEP 1263
Query 336 VTENVVNLYQN 346
VT+NV LY+N
Sbjct 911 VTQNV--LYEN 919
Query 513 SPREGQPVCSQ 523 <<<human integrin beta3
SPR V SQ <<<IDENTITIES
Sbjct 680 SPRRARSVASQ 690 <<<Spike2020 FURIN site, again!!!
Query 338 ENVVNLYQNYSELIPGTTVGVLSMDSSNVLQLIVDAYGKIRSKVELEVR 386
++VVN QN L T V LS + + + D ++ KVE EV+
Sbjct 949 QDVVN--QNAQAL--NTLVKQLSSNFGAISSVLNDILSRL-DKVEAEVQ 992
Query 627 CPTCPDACTFKKECVECKKFD 647
C +C C C C KFD
Sbjct 1240 CCSCLKGCC---SCGSCCKFD 1257
Query 674 TGKDAVNCTYKNEDD---CVV 691
TGK A + YK DD CV+
Sbjct 415 TGKIA-DYNYKLPDDFTGCVI 434
Query 585 NCTTRT 590
N TTRT
Sbjct 17 NLTTRT 22
Query 328 KNINLIFAVTENVVNLYQ----------NYSELIP 352
KN +FA V +Y+ N+S+++P
Sbjct 776 KNTQEVFA---QVKQIYKTPPIKDFGGFNFSQILP 807
Query 334 FAVTENVVNLYQNYSELIPGTTVGVLSMDSSNVLQLIVD 372
FA TE N+ + + I GTT +DS LIV+
Sbjct 92 FASTEKS-NIIRGW---IFGTT-----LDSKTQSLLIVN 121The above is extremely disturbing and serious! So how about the Spike comparison with the very same Integrin alpha-4/beta-7 from Fauci’s patent (https://www.uniprot.org/uniprotkb/P26010/entry)? Here few comparisons:
Query 686 GWCKERTLDNQLFFFLVEDDARGTVV 711 Integrin in Fauci's patent
GW TLD++ L+ ++A V+
Sbjct 103 GWIFGTTLDSKTQSLLIVNNATNVVI 128 SPike2020
over 80 amino acid long entire segment:
Query 2 VALPMVLVL-----LLVLSRGESELDAKIPSTGDATEWRNPHLSMLGSCQPAPSCQKCIL
+A+P + +L +S ++ +D + GD+TE C +L
Sbjct 712 IAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTE-----------------CSNLLL
Query 57 SHPSCAWCKQLNFTASGEAEARRCARREELLAR 89
+ S +C QLN +G A + +E+ A+
Sbjct 755 QYGS--FCTQLNRALTGIA-VEQDKNTQEVFAQ 784
Query 278 RNVSRLLVFTSDDTF 292
RN + T+D+TF
Sbjct 1107 RNFYEPQIITTDNTF 1121
Query 602 MDSCISPEGGLCSGHGRCK 620
M SC S G CS CK
Sbjct 1237 MTSCCSCLKGCCSCGSCCK 1255
Query 503 APGRLGRLCECSVAELSSPDLESGC 527
APG+ G++ + + PD +GC
Sbjct 411 APGQTGKIADYNY---KLPDDFTGC 432
Query 572 ILCGGFGRCQC--GVCHCHANRTGRACECSGD 601
+LC C C G C C G C+ D
Sbjct 1233 MLCCMTSCCSCLKGCCSC-----GSCCKFDED 1259
or the above with better shifting
Query 536 LCSGKGHCQCGRCSCSGQSSGHLCECDDASCE 567
LC C C + C S G C+ D+ E
Sbjct 1234 LCCMTSCCSCLKGCC---SCGSCCKFDEDDSE 1262
Query 366 SAVGELSEDSSNVVQLIMDAYNSLSS 391
SA+G+L + + Q + LSS
Sbjct 943 SALGKLQDVVNQNAQALNTLVKQLSS 968
Query 193 FGSFVDKTVLPFVSTVPSK 211
FG F +LP S PSK
Sbjct 797 FGGFNFSQILPDPSK-PSK 814
Query 157 LMDLS------YSMKDDLERVRQLGHAL---LVRLQEV 185
L D+S +++ +++R+ ++ L L+ LQE+
Sbjct 1166 LGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL 1203
Query 251 VSGNLD 256
VSGN D
Sbjct 1122 VSGNCD 1127
and also the biggest fragment covering the Spike's ACE2 binding domain:
Query 695 NQLFFFLVEDDA---RGTVVLRVRPQEKG--ADHTQAIVLGCVGGIVA---------VGL
N L F V D+ RG V ++ P + G AD+ + G ++A VG
Sbjct 388 NDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGG
Query 741 GLVLAYRLSVEIYDRREYSRFEKEQQQLNWKQDSNP 776
YRL + + FE++ ++ S P
Sbjct 448 NYNYLYRL----FRKSNLKPFERDISTEIYQAGSTP 479Both integrins P05106 and P26010 are related, belong to the same family of proteins(that’s why the name), 40% identity and a BLAST score:
568 bits(1465) 0.0 304/757(40%) 434/757(57%) 31/757(4%)
Query 50 SCQKCILSHPSCAWCKQLNFTASGEAEARRCARREELLARGCPLEELEEPRGQQEVLQDQ
SCQ+C+ P CAWC RC +E LL C E +E P + VL+D+
Sbjct 38 SCQQCLAVSPMCAWCSDEALPLGSP----RCDLKENLLKDNCAPESIEFPVSEARVLEDR
Query 110 PLSQGARGEGA--TQLAPQRVRVTLRPGEPQQLQVRFLRAEGYPVDLYYLMDLSYSMKDD
PLS G+ + TQ++PQR+ + LRP + + ++ + E YPVD+YYLMDLSYSMKDD
Sbjct 94 PLSDKGSGDSSQVTQVSPQRIALRLRPDDSKNFSIQVRQVEDYPVDIYYLMDLSYSMKDD To speed up the detective work, let’s stick with official scientific titles and associations.. When we read “HIV-1 Tat Protein Enters Dysfunctional Endothelial Cells via Integrins and Renders Them Permissive to Virus Replication“ (https://pubmed.ncbi.nlm.nih.gov/33396807/)
and then
“SARS-CoV-2 Mediated Endothelial Dysfunction: The Potential Role of Chronic Oxidative Stress“ (https://pubmed.ncbi.nlm.nih.gov/33519510/)
then we would need to ask, do integrins have anything to do with the known ACE2 affinity for Sars-Cov-2? Only recently (https://pubmed.ncbi.nlm.nih.gov/35150743/) new admission states that:
”Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2”.
And then we get 2012 paper (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3327712/) :
”Angiotensin Converting Enzyme (ACE) and ACE2 Bind Integrins and ACE2 Regulates Integrin Signalling”…
With a quote from it:”These results suggest that ACE2 plays a role in cell-cell interactions, possibly acting to fine-tune integrin signalling. Hence the expression and cleavage of ACE2 at the plasma membrane may influence cell-extracellular matrix interactions and the signalling that mediates cell survival and proliferation. As such, ectodomain shedding of ACE2 may play a role in the process of pathological cardiac remodelling.”
So if the first HIV paper states: “Tat protein of Human Immunodeficiency Virus (HIV)-1 is released by acutely infected cells in a biologically active form and enters dendritic cells upon the binding of its arginine-glycine-aspartic acid (RGD) domain to the α5β1, αvβ3, and αvβ5 integrins.“ and then we suddenly get the very same binding RGD motif in the very first ‘NEW’ SARS-CoV-2 Spike, sharing the extremely unique cystein cluster, part of the furin site, with that ‘RGD’not even present in Spikes of OLD coronaviruses, shall we believe that, all that is random???
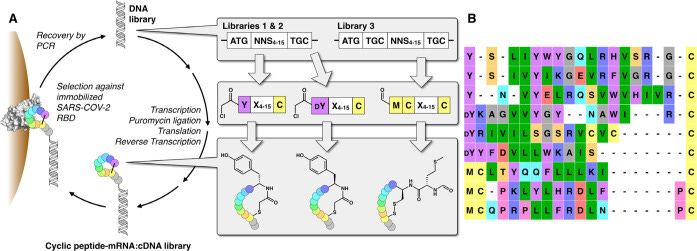
Is that why Fauci, the HIV expert warned in 2017 Of ‘Surprise Outbreak’ During Trump Administration, a YEAR BEFORE he got his patent??? Fauci’s solution to HIV was: “In several examples, the α4 integrin antagonist is a monoclonal antibody that specifically binds to a α4, β1 or β7 integrin subunit or a cyclic hexapeptide with the amino acid sequence of CWLDVC.“ Here most recent research on: “Discovery of Cyclic Peptide Ligands to the SARS-CoV-2 Spike Protein Using mRNA Display” with a picture displaying the sequences:
The above cyclic peptides, containig the terminal cysteins, bind the receptor binding domain of ACE2 on Spike2020, Fauci’s cyclic peptide binds to the integrins!! Arent’ they all sharing the above similarities?? Does Fauci know all this? If something needs to be ‘fact-checked’ involving BIG names it almost instantaneously implies: check it once again! Just to show once again, how many times Fauci’s octapeptide CxxxxC aligns with the C-terminal part of the Spike, injected into billions:
Sbjct 1235 CCMTSCCSCLKGCCSCGSCCK 1244 SPIKE 2020
Query 2 CWLDVC 8 Fauci's patented cyclic 'HIV-octapeptide'
Query 3 CWLDVC 8
Query 4 CWLDVC 8
Query 5 CWLDVC 8 One of the very kind readers, Sally Gold, pointed out in the comment section to another extremely important fact I didn’t know, namely that Sars-Cov2 infection depends stronly on the expression level of cathepsin peptidase^9. That cysteine protease according to MEROPS database, contributes to the generation of anigenic peptides for the MHC II system. A licensed anti-influenza drug, amantadine, inhibiting that protease, prevented SARS-CoV-2 infection. Also Teicoplanin^10, an antibacterial drug previously used to treat virus infections caused by influenza virus, hepatitis C virus, HIV, Ebola, flavivirus, as well as MERS-CoV and SARS-CoV, works equally for SARS-CoV-2. Teicoplanin acts as a CatL inhibitor, blocking the S protein cleavage (Zhou et al., 2016; Yousefi et al., 2020). Doesn’t is sound that one and the same drug is capable of inhibiting ALL THE Spikes of all the most infectious diseases??? What does the cathepsin L does in HIV infection? “HIV Infection Induces Extracellular Cathepsin B Uptake and Damage to Neurons“^12, exactly like SARS-CoV-2 Spike. And if “HIV Protease Inhibitor-Induced Cathepsin Modulation Alters Antigen Processing and Cross-Presentation”^12 and this process by which pathogen Ags are internalized, degraded, and presented by MHC class I, is crucial to prime CD8 T cell responses, than that’s exactly the same what happens in SARS-CoV-2. BUT if the degardation pieces are very similar, due to high homology of all the sequences between the Spikes, then we’ll end up with similar drugs blocking the same targeting family of proteases.. Last example “Cathepsins and Their Endogenous Inhibitors in Host Defense During Mycobacterium tuberculosis and HIV Infection“ ^13 with its quote:”The moment a very old bacterial pathogen met a young virus from the 80’s defined the beginning of a tragic syndemic for humanity.” indicates all these common pathways leading to specific dis-eases, where one and the same immune system consisting of the very same T cells, B cells, CD4, CD8, etc. has to deal with an EXTERNAL foreign intruder, clearly always RECOGNIZED as such. NO MORE NOW, with covid genetically modifying injections. NOW Humans are the intruders themselves, literally, by expressing the very pathogenic injected agent they should avoid!
The main point of these comparisons is to point out the fact, that in order to propagate for example viral SARS-CoV-2 infection with a big pool of identical building blocks common with HIV-1, with Marburg, with all the other viruses mentioned in previous posts, incredible large portions of HUMAN proteins, any genetic re-programming of the human body for synthetic SARS Spikes production will automatically affect infections with other even more lethal or maybe even benign viruses.
Craig Venter, the ‘genetics genius’, expressed in few simple examples^13 the importance of the NATURAL mRNA content of every single cell:
-”CCR5 mRNA is significantly increased in a woman with STDs (sexually transmitted diseases) and inflammatory conditions, so the bacterial environment in conjunction with a woman’s genetics could determine her susceptibility to HIV.”^13
-in vivo “knockdown” of TRIM30a mRNA by small interfering RNA impaired LPS-induced tolerance.
-Intracellular components create myriad metabolites that can interfere and alter the
correct transcription of human proteins. Some of these metabolites can also disrupt
cellular repair mechanisms, resulting in the accumulation of “junk” (e.g., proteins,
enzymes, and mRNA) in the cytosol.
With one word, the mRNA status in every cell mirrors its health, or disease. Venter investigated mRNA profiling of thousands of traditionally HEP vaccinated people, in 2020 (right on time), all realted to human disease, and yet, Fauci didn’t seem to have from him the same information on Spike mod mRNA before changing that profiling in billions of covid injected HUMAN victims starting on global scale in 2021!?
The oldest protein in which I can find traces related to Spike, carrying actully the furin site, just shifted in the sequence, is the s.c. peplomeric protein, filed for a patent in 1986 (https://www.ncbi.nlm.nih.gov/nuccore/A24863.1). The alignment of the 2020 SPike and the 1986 furin site from the Infectious bronchitis virus spike looks following:
20.4 bits(41) 6e-04 Compositional matrix adjust. 12/34(35%) 15/34(44%) 1/34(2%)
Query 22 TTPSSPRR-RSFIEDLLFTSVESVGLPTDDAYKN 54 Spike 1986 furin site
T +SPRR RS + S+G AY N
Sbjct 25 TQTNSPRRARSVASQSIIAYTMSLGAENSVAYSN 58 Spike 2020 furin site
Patent: EP 0221609-A1 13-MAY-1987; DUPHAR INTERNATIONAL RESEARCH B.V
The score of the alignment of the entire 1986 Spike with the 2020 Spike, with 100% overlap and ~37% identities is 364, here just first 3 raws of the alignment:
364 bits(935) 7e-111 Compositional matrix adjust. 202/548(37%) 299/548(54%) 22/548(4%)
NVLIPNSFNLTVTDEYIQTRMDKVQINCLQYVCGNSLDCRDLFQQYGPVCDNILSVVNSI 642 1987
++ IP +F ++VT E + M K ++C Y+CG+S +C +L QYG C + + I
SIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGI 770 2020
GQKEDMELLNFYSSTKPAGFNTPFLSNVSTGEFNISLLLTTPSSPRRRSFIEDLLFTSVE 702 1987
++D ++ K + TP + + G FN S +L PS P +RSFIEDLLF V
AVEQDKNTQEVFAQVKQI-YKTPPIKDF--GGFNFSQILPDPSKPSKRSFIEDLLFNKVT 827 2020
SVGLPTDDAYKNCTAGPLGFLKDLACAREYNGLLVLPPIITAEMQTLYTSSLVASMAFGG 762 1987
Y +C +DL CA+++NGL VLPP++T EM YTS+L+A G
LADAGFIKQYGDCLGDIAA--RDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSG 885 2020Few clarifications: the entire SARS-CoV-2 3D model exists as a genetic sequence, Pdb Data Bank does NOT have SARS-CoV-2 three dimentional structure of all componds (usually determined by X-Ray crystallography or Electron Microscopy). The OFFICIALLY known SARS-CoV-2 3D parts are some of the viral sub-components, like the Spike, with all its mutations.
For those who claim all is just graphene, frequencies, I have this counterargument: molecular biologists, geneticists created your GMO food, where mixed species (bugs+viruses+plants) were produced and served on people’s tables withut their consent, for the last ~25 YEARS! Do you think they will stop doing the same with the entire human body with incredible wonderful properties?? If you apply JUST graphene to a mixture of proteins/DNA/RNA you get a RANDOM cutting events without too much intelligence behind (my believe). YOU NEED THE WONDER of being a HUMAN and having a great HEART and BRAIN in order to function as such. In order to negate all the HUMAN BEAUTY, you need to be Dr. Fauci. Here a quote from him from the ‘Real Anthony Fauci.“ Part2:
“The worst bioterrorist is nature itself. The chances of Nature creating something really bad is much better the we near mortal humans doing it” « what a bunch of crap!!!
Dr. Fauci, look into the mirror and find the nature’s error in your creation.
For those interested, Optogenetics will continue.. Right now, FAuCI comes first.
LITERATURE.
Josephine C Adams et al. “The evolution of tenascins and fibronectin“ Cell Adh Migr. 2015;9(1-2):22-33. https://pubmed.ncbi.nlm.nih.gov/25482621/
Sylvia Fitting et al. Brain Res. 2008 September 26; 1232: 139–154.
Sylvia Fitting et. al. Curr HIV Res. 2015; 13(1): 21–42. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4467793/
Kathy Lien et al. PLoS One. 2019; 14(12): e0226343. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6927651/
Junyoung Oh et al. Sci Rep. 2022 Mar 31;12(1):5496. https://pubmed.ncbi.nlm.nih.gov/35361832/
Shuibing Chen et al. Res Sq. 2021 May 21;rs.3.rs-513461. https://pubmed.ncbi.nlm.nih.gov/34031650/
Yun-Ju Lai et al. Am J Cancer Res. 2021 May 15;11(5):2278-2290. https://pubmed.ncbi.nlm.nih.gov/34094684/
https://academictree.org/chemistry/publications.php?pid=52415
with following titles:Molteni A, Zakheim RM, Mullis KB, Mattioli L. The effect of chronic alveolar hypoxia on lung and serum angiotensin I converting enzyme activity. Proceedings of the Society For Experimental Biology and Medicine. Society For Experimental Biology and Medicine (New York, N.Y.). 147: 263-5. PMID 4373751
Zakheim RM, Molteni A, Mattioli L, Mullis KB. Angiotensin I-converting enzyme and angiotensin II levels in women receiving an oral contraceptive. The Journal of Clinical Endocrinology and Metabolism. 42: 588-9. PMID 176173 DOI: 10.1210/Jcem-42-3-588
Mattioli L, Zakheim RM, Mullis K, Molteni A. Angiotensin-I-converting enzyme activity in idiopathic respiratory distress syndrome of the newborn infant and in experimental alveolar hypoxia in mice. The Journal of Pediatrics. 87: 97-101. PMID 168336
Molteni A, Mullis KB, Zakheim RM, Mattioli L. The effect of changes in dietary sodium on lung and serum angiotensin I converting enzyme in the rat Laboratory Investigation. 35: 569-573. PMID 186664
Zakheim RM, Mattioli L, Molteni A, Mullis KB, Bartley J. Prevention of pulmonary vascular changes of chronic alveolar hypoxia by inhibition of angiotensin I-converting enzyme in the rat. Laboratory Investigation; a Journal of Technical Methods and Pathology. 33: 57-61. PMID 167232
Miao-Miao Zhao et al. “Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development“ Signal Transduct Target Ther. 2021 Mar 27;6(1):134.
Caio P. Gomes et al. “Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics” Front Cell Infect Microbiol. 2020; 10: 589505.
Published online 2020 Dec 8. doi: 10.3389/fcimb.2020.589505
Yisel M Cantres-Rosario et al. Sci Rep. 2019 May 29;9(1):8006. https://pubmed.ncbi.nlm.nih.gov/31142756/
Georgio Kourjianet al. J Immunol. 2016 May 1;196(9):3595-607. https://pubmed.ncbi.nlm.nih.gov/27009491/
“Metagenomics of the Human Body” by Craig VEnter





Bravo to the few of european parliament members. Ursula should not just resign, she should be investigated and punished. Otherwise another one like her will be happy to take her place and continue to do the same crimes and abuses. We can expect anything when the corporations powers and money are above the law and governments so here we are, abused, agresed, put at the corner and subjects of their criminal experiments.
Where you said, "very unique cysteine cluster at the C-terinal of Spike", I assume you meant C-terminal instead of C-terinal. If so, it may be worth correcting.